
Needleman wunsch algorithm python serial#
Moreover, the computation time of the proposed heterogeneous anti-diagonal approach increases gradually despite the big increments in sequence length, whereas the computation time of the serial approach grows rapidly.īiological pairwise sequence alignment can be used as a method for arranging two biological sequence characters to identify regions of similarity. The experiment showed that the proposed approach outperforms the serial method in terms of computation time by approximately three times. Measurements of computation times are performed under the same experimental setup and using various pairwise sequences at different lengths. We then measure and compare the computation time between the proposed approach and a straightforward serial approach that uses CPU only. As a solution, this research introduces the heterogeneous anti-diagonal approach, which benefits from the interaction between the serial implementation on CPU and the parallel implementation on GPU. However, there is a data dependency issue due to the property of a dynamic programming algorithm. This research aims to parallelize the computation involved in the algorithm to speed up the performance using CUDA.
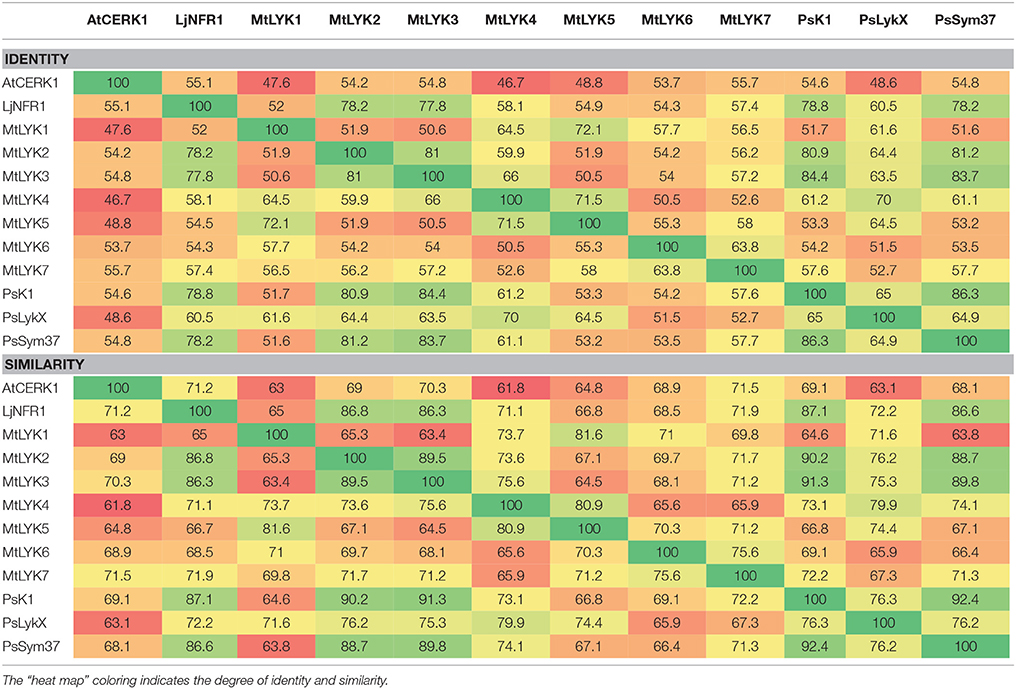
The task becomes nontrivial as the number of sequences to compare or the length of sequences increases. Needleman-Wunsch dynamic programming algorithm measures the similarity of the pairwise sequence and finds the optimal pair given the number of sequences.


 0 kommentar(er)
0 kommentar(er)
